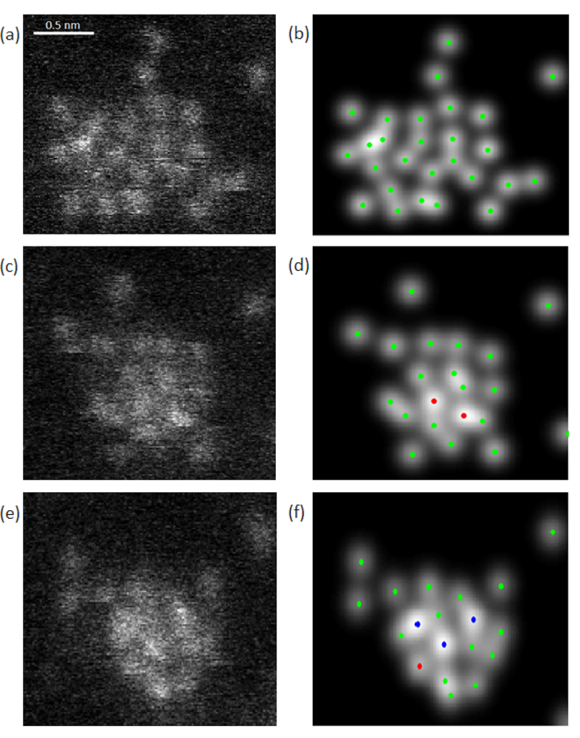
(Oct 2022) We introduce a Bayesian genetic algorithm for reconstructing atomic models of monotype crystalline nanoparticles from a single projection using Z-contrast imaging. The results show promising prospects for obtaining reliable reconstructions of beam-sensitive nanoparticles during dynamical processes from images acquired with sufficiently low incident electron doses. This work was published in npj Computational Materials (“Experimental reconstructions of 3D atomic structures from electron microscopy images using a Bayesian genetic algorithm“)
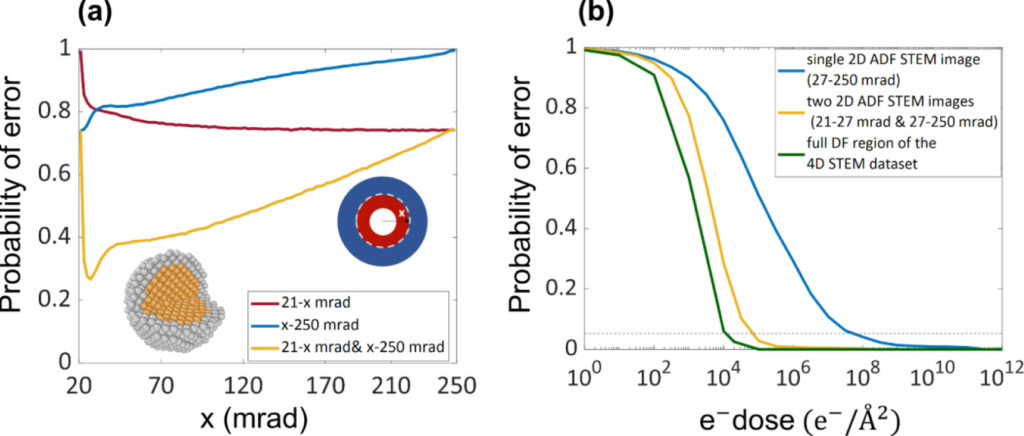
(Oct 2022) In this work we proposed an optimal experiment design for element specific atom-counting. For this purpose the probability of error is quantified and the benefit of multiple STEM detectors is investigated. The approach was tested on a wide variety of possible heterogeneous configurations. This work was published in Ultramicroscopy (“Optimal experiment design for element specific atom counting using multiple annular dark field scanning transmission electron microscopy detectors“)
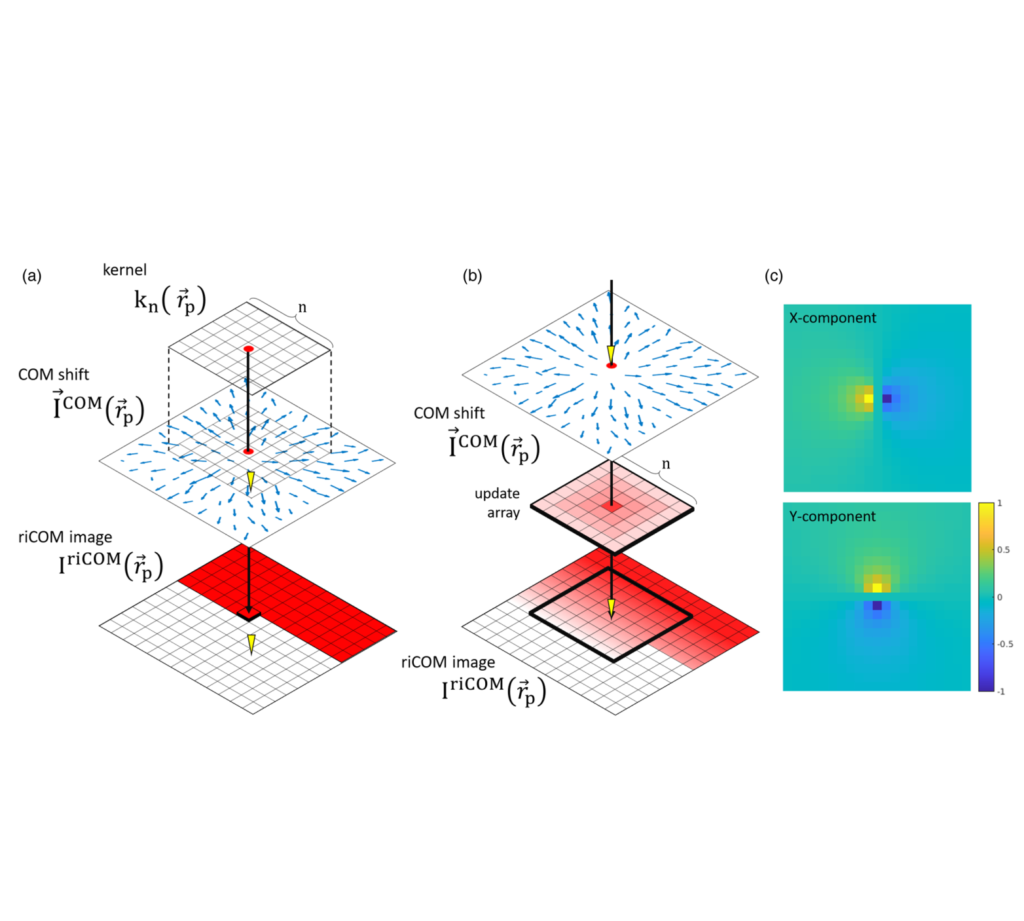
(Apr 2022) We proposed a real-time image reconstruction method for scanning transmission electron microscopy (STEM). With an algorithm requiring only the center of mass of the diffraction pattern at one probe position at a time, it is able to update the resulting image each time a new probe position is visited without storing any intermediate diffraction patterns. Compared with other reconstruction methods, the proposed method produces high-quality reconstructions with good noise robustness at extremely low memory and computational requirements. An efficient, interactive open source implementation of the concept is further presented, which is compatible with frame-based, as well as event-based camera/file types. This work was published in Microscopy and Microanalysis (“Real-Time Integration Center of Mass (riCOM) Reconstruction for 4D STEM“)
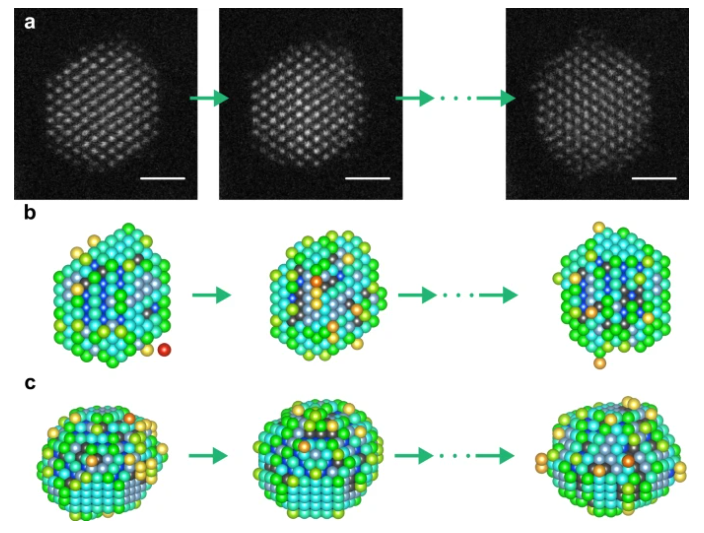
(Nov 2021) We made it possible to investigate the 3D atomic structure of supported NPs at high temperatures, which may deviate from their ground state configuration. In order to achieve this advancement, we proposed an optimized approach by using an iterative local minima search algorithm followed by molecular dynamics structural relaxation of candidate structures associated with each local minimum. This work was published in Small Methods (“3D Atomic Structure of Supported Metallic Nanoparticles Estimated from 2D ADF STEM Images: A Combination of Atom-Counting and a Local Minima Search Algorithm“) and Nanoscale (“Three-dimensional atomic structure of supported Au nanoparticles at high temperature“). The Open Access version of the papers can be accessed here and here.
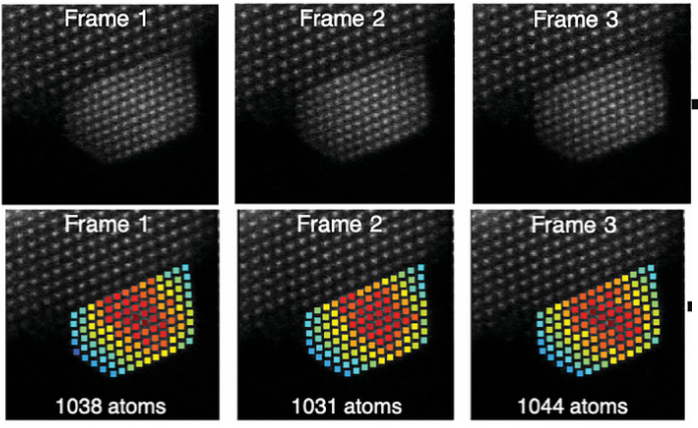
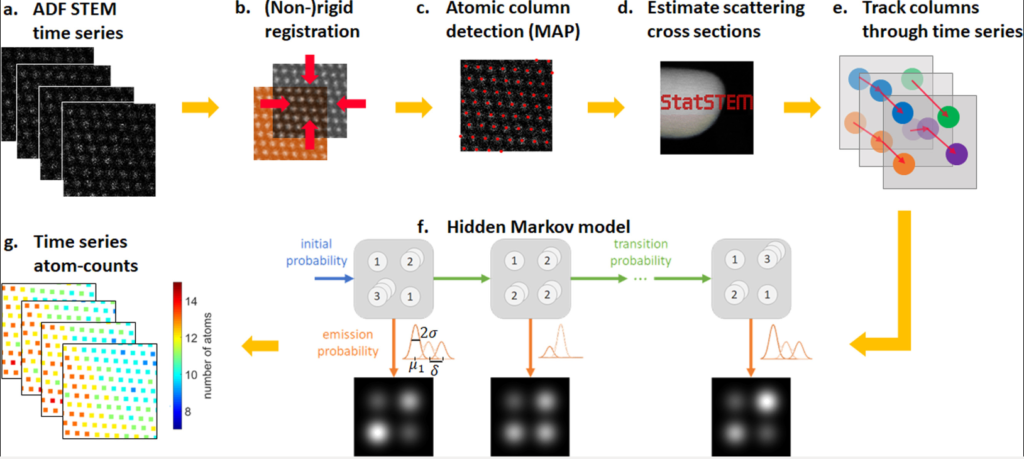
(Oct 2021) We introduced hidden Markov models to quantify sequential ADF STEM images and used them for atom-counting from time series data resulting in reliable quantitative analyses of atomic scale dynamic processes. This paper was published in Ultramicroscopy (“Hidden Markov model for atom-counting from sequential ADF STEM images: Methodology, possibilities and limitations“). The Open Access version of the paper can be access here.
(Mar 2020) A dynamic game of hide and seek at the nanoscale (Nanowerk news item). We propose a new method to measure atomic scale dynamics of nanoparticles from experimental high-resolution annular dark field scanning transmission electron microscopy images. By using the so-called hidden Markov model, which explicitly models the possibility of structural changes, the number of atoms in each atomic column can be quantified over time. This work was published in Physical Review Letters (“Measuring Dynamic Structural Changes of Nanoparticles at the Atomic Scale Using Scanning Transmission Electron Microscopy”).
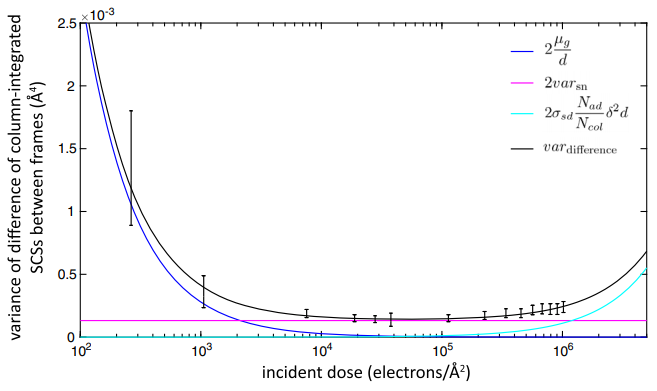
(Feb 2019) We have developed and experimentally validated a theoretical model that can be used to predict the optimal electron dose to quantify unknown nanostructures in their native state with the highest precision before conducting the experiment. “Control of Knock-On Damage for 3D Atomic Scale Quantification of Nanostructures: Making Every Electron Count in Scanning Transmission Electron Microscopy”. Van Aert S, De Backer A, Jones L, Martinez GT, Béché, A, Nellist PD, Physical Review Letters 122, 066101 (2019). (link to Open Access pdf)
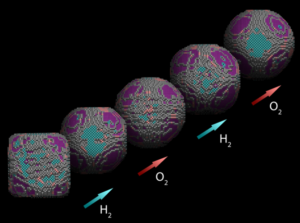
(Jan 2019) We have contributed to the investigation of nanoparticles in a gaseous environment by combining atom counting and 3D modelling from a single viewing direction.
“Three-Dimensional Quantification of the Facet Evolution of Pt Nanoparticles in a Variable Gaseous Environment”, Altantzis T., Lobato I., De Backer A., Béché A., Zhang Y., Basak S., Porcu M., Xu Q., Sánchez-Iglesias A., Liz-Marzán L.M., Van Tendeloo G., Van Aert S., Bals S.
Nano Lett 19, 477 (2019). (link to Open Access pdf)
(Aug 2018) Statistical model removes human bias in finding individual atoms in fuzzy images. A statistical model, developed by researchers from Belgium, the Netherlands and the UK, promises to resolve fuzzy TEM images down to single atoms – even light ones that only weakly scatter electrons and are therefore hard to spot. At the same time, the method removes human bias from the equation.
Chemistry World science blog
“Single Atom Detection from Low Contrast-to-Noise Ratio Electron Microscopy Images”. Fatermans J, den Dekker A J, Müller-Caspary K, Lobato I, O’Leary C M, Nellist P D, Van Aert S, Physical Review Letters 121, 056101 (2018). (link to Open Access pdf)